The following is a short list of my hero in this field (no particular order, I just randomly think who I will add here), it may different from yours, or even missing some big figures. But remember, this is my list!
Geoff Hinton, from University of Toronto, who is one of the main force to push neural network, and deep learning forward. I especially love this video - The Deep Learning Saga
Michael Jordan from UC Berkeley. Just look at the list of his past students and postdocs, you will realize how many great experts he taught before! I took his class before, but at that moment, I can not understand all!
Trevor Hastie and Robert Tibshirani I learned most of my machine learning knowledge from their books - An introduction to Statistical Learning and The Elements of Statistical Learning. If you don't know who invented Lasso method we just talked above, google it!
Andrew Ng, from Stanford. Most people knows Andrew probably from the Coursera machine learning course. Recently, he is very active in machine learning field.
Tom M. Mitchell, from Carnegie Mellon University. I first know him is from his book machine learning, a very readable introduction.
Yann LeCun, another driving force in deep learning. He is best known for his contribution in the convulutional neural networks.
Christopher Bishop, from University of Edinburdh. He was trained as a physicists, but then later became an expert in machine learning. His book Pattern Recognition and Machine Learning is another must have if you want to learn machine learning.
Yaser S. Abu-Mostafa, Caltech. I took his learning from data online, and it is my first step in machine learning!
Jake VanderPlas, University of Washington. I used so many tutorials and slides from him!
This lists will be updated in the future ^)^
Sunday, August 28, 2016
Wednesday, August 17, 2016
PyData San Francisco 2016
I attended PyData San Francisco 2016. It is a nice short 2 day conference, even though I only made for the first day. Here are also some interesting things I learned from this conference. Also, this is my lightning talk video.
Packages
The Journal of Open Source Software
A developer friendly journal for research software packages. It is a nice place to publish your open source software for credits.
A developer friendly journal for research software packages. It is a nice place to publish your open source software for credits.
SnakeViz
A browser based graphical viewer for the output of Python’s cProfile module
A browser based graphical viewer for the output of Python’s cProfile module
digdag
A simple tool that helps you to build, run, schedule, and monitor complex pipelines of tasks. It handles dependency resolution so that tasks run in order or in parallel.
A simple tool that helps you to build, run, schedule, and monitor complex pipelines of tasks. It handles dependency resolution so that tasks run in order or in parallel.
airflow
A platform to programmatically author, schedule and monitor workflows.
A platform to programmatically author, schedule and monitor workflows.
nipype
A framework for developing efficient and reliable medical imaging data analysis pipelines.
A framework for developing efficient and reliable medical imaging data analysis pipelines.
PyFlux
A time series library for the Python programming language.
A time series library for the Python programming language.
Carousel
Model Simulation Framework
Model Simulation Framework
ParseHub
Easily extract data from any website.
Easily extract data from any website.
Docplex
With this library, you can quickly and easily add the power of optimization to your application.
With this library, you can quickly and easily add the power of optimization to your application.
3D XPoints
The next generation of storage that will revolutionize computing.
The next generation of storage that will revolutionize computing.
The video of the talks are still not available. I will link some of the interesting talks from my point of view here when they are online.
Labels:
conference,
data science,
python
Location:
San Francisco, CA, USA
Friday, August 12, 2016
Sentinel-1A data processing using GMTSAR
I learned processing InSAR data in the short course at Scripps using the GMTSAR. In this blog, I will write down the steps I used to process the Sentinel-1A data for my own reference. I will use the 2016 Taiwan M6.4 Meinong earthquake as an example. You can find all the files and figures at Qingkai's Github. Most of the content is written during the short course by the help of the educators.
Step 1 - get data
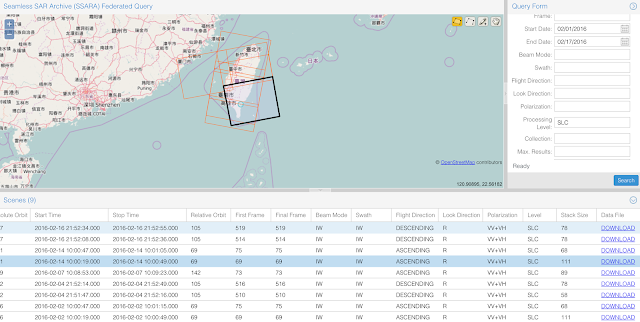
There are multiple ways to get the data. The most common way is to use the GUI or use the API directly. For this earthquake, we will try to use the GUI to download the Sentinel-1A data. We first draw a polygon around the earthquake region as shown in the following figure. Since the earthquake occurred on Feb 5th, we put our search range from Feb 1 - Feb 17. Because we only use the SLC processing level, so I changed the 'Processing Level' to 'SLC'.
After you pressed the search button, you will get a list of available data within the range, shown below. When you select any of the record, it will highlight on the map as the black box. Scroll to the right, you will see the download button two download the data. Because we want to download the data that can reflect the deformation caused by the earthquake, therefore, we need both the data before the earthquake, and after. By looking at the Ascending path, we have Feb 2nd and Feb 14th satisfy the requirement (Note: You should choose both from the same path, either Ascending or Descending). Then go ahead to download the data (about 10 Gb).
Step 2 - get the orbit data
You also need the orbits data before processing, which you can download it here. The naming of the orbits data is like
S1A_OPER_AUX_POEORB_OPOD_20160222T121629_V20160201T225943_20160203T005943.EOF
Note there are 3 dates, the first one indicate when it is processed (mostly useless in our example), and the last two dates are the start and end dates of the orbits, and we need make sure our Sentinel data date is within this range. For our case, we need the following two files:
S1A_OPER_AUX_POEORB_OPOD_20160222T121629_V20160201T225943_20160203T005943.EOF
S1A_OPER_AUX_POEORB_OPOD_20160305T121418_V20160213T225943_20160215T005943.EOFStep 3 - folder structure
The next step you need is to put the data into a folder structure for processing. The folder structure looks like the following, and the red color indicate directories, you can see the subdirectory or the files in them. The two .SAFE directories are from the InSAR data you downloaded. And the two files ending with eof.txt are the orbits data we downloaded. For the dem.grd, you can download it from here by specify the latitude and longitude. The 01_run_prep.sh, 02_run_proc.sh, and config.s1a.txt, we will talk in the next section.
Taiwan_earthquake ├── 01_run_prep.sh ├── 02_run_proc.sh ├── config.s1a.txt ├── orbits | ├── S1A_IW_SLC__1SDV_20160202T100019_20160202T100049_009766_00E469_C190.SAFE | ├── S1A_IW_SLC__1SDV_20160214T100019_20160214T100049_009941_00E981_ABD9.SAFE | ├── S1A_OPER_AUX_POEORB_OPOD_20160222T121629_V20160201T225943_20160203T005943.eof.txt | └── S1A_OPER_AUX_POEORB_OPOD_20160305T121418_V20160213T225943_20160215T005943.eof.txt └── topo └── dem.grd
Step 4 - change the config file
Before you run the following steps, you need consider the settings in the config.s1a.txt file. There are many settings inside, but you can first run with the default setting, and change it later. For more details, you should refer to the GMTSAR documentation. My example config.s1a.txt file is here:
config.s1a.txt
Step 5 - run the preparation
The 01_run_prep.sh script should be run first to prepare the data and align the master and slave images. Before you run that, you need change the name of the files inside. Note that the input of the align_tops_esd.csh run in the script is: InSAR-1, orbit-1, InSAR-2, orbit-2, dem.grd. Notice that, I only use the polarity 'vv' for this example. See my example file:
01_run_prep.sh.
After you run this script, you will find you have a updated folder structure, which contains 4 more folders: F1, F2, F3, and raw. The F1, F2, and F3 folder contain the aligned data for the 3 subswaths.
Taiwan_earthquake ├── 01_run_prep.sh ├── 02_run_proc.sh ├── config.s1a.txt ├── F1 | ├── raw | ├── topo | └── config.s1a.txt ├── F2 | ├── raw | ├── topo | └── config.s1a.txt ├── F3 | ├── raw | ├── topo | └── config.s1a.txt ├── raw | └── many things here ^_^ ├── orbits | ├── S1A_IW_SLC__1SDV_20160202T100019_20160202T100049_009766_00E469_C190.SAFE | ├── S1A_IW_SLC__1SDV_20160214T100019_20160214T100049_009941_00E981_ABD9.SAFE | ├── S1A_OPER_AUX_POEORB_OPOD_20160222T121629_V20160201T225943_20160203T005943.eof.txt | └── S1A_OPER_AUX_POEORB_OPOD_20160305T121418_V20160213T225943_20160215T005943.eof.txt └── topo └── dem.grd
Step 6 - run the processing
Now, if everything works fine, you can step into the last step: run the processing script - 02_run_proc.sh, again, you need change the name of the files inside the script, and this one is easier. You can find my example file here:
02_run_proc.sh.
Results
After all the above steps, you will find in each of the subswath folder (i.e., F1, F2, and F3), there are two more folders: intf, SLC. Most of the results figures/data are stored in the intf folder. Here are my results:
wrapped fringes:
wrapped fringes:
unwrapped:
The next step you want to do is to remove the trend from the unwrapped image.
Acknowledgement
Thank:
UNAVCO for providing the financial support.
GMTSAR developer team for the great workshop.
GMT team for great support!
UNAVCO for providing the financial support.
GMTSAR developer team for the great workshop.
GMT team for great support!
Wednesday, August 10, 2016
Visiting University of California, San Diego
I am currently visiting University of California at San Diego. The campus is really beautiful, I will show some of the pictures below when I was wondering in the campus.
Eucalyptus trees
Before I die
Geisel Library
Art work
Sun God and King Triton
Eucalyptus trees
Before I die
Geisel Library
Art work
View from Scripps
Sunday, August 7, 2016
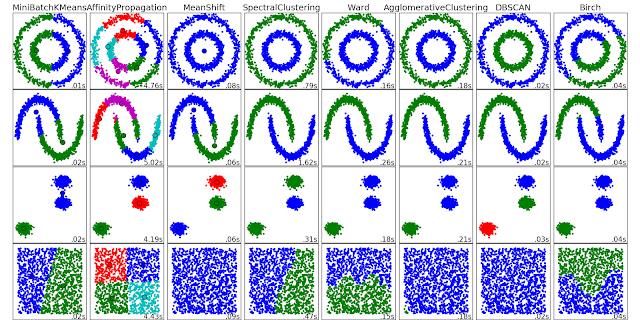
Clustering with DBSCAN
I am currently checking out a clustering algorithm: DBSCAN (Density-Based Spatial Clustering of Application with Noise). As the name suggested, it is a density based clustering algorithm: given a set of points in some space, it groups together points that are closely packed together (points with many nearby neighbors), and marks points as outliers if they lie alone in low-density regions. It has many advantages, including no need specify the number of clusters, can find arbitrary shaped clusters, relatively fast, etc. Of course, there's no single algorithm can do everything, DBSCAN has disadvantage as well.
The algorithm has two parameters: epsilon and min_samples, the advantage of the algorithm is that you don’t have to specify how many clusters you need, it can find all the clusters that satisfy the requirement. For the disadvantage, it is very sensitive to the parameter you choose.
The summary of this algorithm is:
Step 1: For each point in the dataset, we draw a n-dimensional sphere of radius epsilon around the point (if you have n-dimensional data).
Step 2: If the number of points inside the sphere is larger than min_samples, we set the center of the sphere as a cluster, and all the points within the sphere are belong to this cluster.
Step 3: Loop through all the points within the sphere with the above 2 steps, and expand the cluster whenever it satisfy the 2 rules.
Step 4: For the points not belong to any cluster, you can ignore them, or treat them as outliers.
Step 1: For each point in the dataset, we draw a n-dimensional sphere of radius epsilon around the point (if you have n-dimensional data).
Step 2: If the number of points inside the sphere is larger than min_samples, we set the center of the sphere as a cluster, and all the points within the sphere are belong to this cluster.
Step 3: Loop through all the points within the sphere with the above 2 steps, and expand the cluster whenever it satisfy the 2 rules.
Step 4: For the points not belong to any cluster, you can ignore them, or treat them as outliers.
The original paper about DBSCAN was published 10 years ago in 1996, and can be found here. It is currently ranked the 41st place in the most cited publication in data mining. If you find the paper is too heavy on defining different points, you can check this very nice video on youtube shows how this works: Here. Scikit learn already has a very nice example to show the effectiveness of the algorithm. You can find the example here.
In the following, we will cluster the Loma Prieta earthquake aftershocks using DBSCAN. You can find the code at Qingkai's Github.
Grab the earthquake data
%pylab inline
import pandas as pd
import numpy as np
import folium
from mpl_toolkits.basemap import Basemap
import matplotlib.pyplot as plt
Earthquake data can be get from the ANSS catalog. For simplicity, I just download aftershock data above Magnitude 2.0 for one week after the Loma Prieta earthquake. All together, there are 1009 earthquakes in this region.
# Read in earthquake data
df = pd.read_csv('./LomaPrieta_aftershocks_1week_above_2.csv', skiprows = 7)
# Get the latitude and logitude of the earthquakes
coords = df.as_matrix(columns=['Latitude', 'Longitude'])# Plot the location of the earthquakes
plt.figure(figsize = (12, 12))
m = Basemap(projection='merc', resolution='l', epsg = 4269,
llcrnrlon=-122.7,llcrnrlat=36.2, urcrnrlon=-120.8,urcrnrlat=37.5)
# plot the aftershock
x, y = m(coords[:, 1], coords[:, 0])
m.scatter(x,y,5,marker='o',color='b')
m.arcgisimage(service='World_Shaded_Relief', xpixels = 5000, verbose= False)
plt.show()Cluster and see the results
We are now using the DBSCAN from the sklearn. I followed Geoff Boeing's blog to cluster the geo-spatial data using the metrics haversine. I choose the epsilon roughly 1.5 km, and the min_samples = 5. We can see that DBSCAN detected 9 clusters in different colors (note that the black dots are identified as outliers).
from sklearn.cluster import DBSCAN
from sklearn import metrics
kms_per_radian = 6371.0088
epsilon = 1.5 / kms_per_radian
# Run the DBSCAN from sklearn
db = DBSCAN(eps=epsilon, min_samples=5, algorithm='ball_tree', \
metric='haversine').fit(np.radians(coords))
cluster_labels = db.labels_
n_clusters = len(set(cluster_labels))
# get the cluster
# cluster_labels = -1 means outliers
clusters = \
pd.Series([coords[cluster_labels == n] for n in range(-1, n_clusters)])import matplotlib.cm as cmx
import matplotlib.colors as colors
# define a helper function to get the colors for different clusters
def get_cmap(N):
'''
Returns a function that maps each index in 0, 1, ... N-1 to a distinct
RGB color.
'''
color_norm = colors.Normalize(vmin=0, vmax=N-1)
scalar_map = cmx.ScalarMappable(norm=color_norm, cmap='nipy_spectral')
def map_index_to_rgb_color(index):
return scalar_map.to_rgba(index)
return map_index_to_rgb_colorplt.figure(figsize = (12, 12))
m = Basemap(projection='merc', resolution='l', epsg = 4269,
llcrnrlon=-122.7,llcrnrlat=36.2, urcrnrlon=-120.8,urcrnrlat=37.5)
unique_label = np.unique(cluster_labels)
# get different color for different cluster
cmaps = get_cmap(n_clusters)
# plot different clusters on map, note that the black dots are
# outliers that not belone to any cluster.
for i, cluster in enumerate(clusters):
lons_select = cluster[:, 1]
lats_select = cluster[:, 0]
x, y = m(lons_select, lats_select)
m.scatter(x,y,5,marker='o',color=cmaps(i), zorder = 10)
m.arcgisimage(service='World_Shaded_Relief', xpixels = 5000, verbose= False)
plt.show()
Labels:
Algorithms,
data science,
python
Location:
Berkeley, CA 94702, USA
Subscribe to:
Posts (Atom)